
Figure 2: Example BIO control display
Before using this API, it is important to understand the concept of BIO messaging. A brief introduction was given in the section BIO Messages but more extensive information can be found in the SDK documentation and at Forum Nokia.
Crucial to the use of BIO messaging, is the definition of a BIO Information File (BIF). This defines the UID of the BIO message and contains information about the parser and BIO control to be used for the particular BIO message type. The BIF is created from a resource during the normal build process, that is using the standard resource compiler. An example resource file is shown here:
#include <biftool2.rh>
RESOURCE BIO_INFO_FILE
{
message_type_UID=0x1000FFFF; // Unique UID for the message type
message_parser_name="BIOControlApiParser.dll"; // Parser name (a parser is not necessary for using the plugin control)
message_app_UID=KUidUseDefaultApp; // This defines that the message is opened in the default app (SMS editor)
message_appctrl_name="BIOControlApi.dll"; // this is the name of our own BIOcontrol
description="BioExample"; // Displayed in the Inbox
ids=
{
ID
{
type=ENbs; // Narrow band (SMS)
confidence=EPossible;
character_set=KCharSetDefault;
text="//MYTK"; // Initial characters to define our bio message
}
};
}
The example BIF resource above defines both a BIO parser and a BIO control.
To use Plugin BIO control API, the parser is optional but it is recommended
that a parser should be used, as a minimum, to set the parsed flag to read.
The Symbian Generic File Parser (gfp.dll) can be used for
this purpose.
A polymorphic DLL is created for the BIO control (CMsgBioControl derived
class) and this is referenced in the BIF: message_appctrl_name="BIOControlApi.dll".
Since the control is used as a plug-in to the messaging UI, the message_app_UID should
be defined as KUidUseDefaultApp - that is, the SMS editor.
While it is possible to define a specific port number for BIO messages
to be received on, this example just uses a simple text string as an identifier.
The type ENbs indicates that, in this instance, the bearer
is SMS. The string is written as the first characters of the BIO message.
On the target device, the BIF file created should reside in the directory \resource\messaging\bif (The parser DLL and control DLL both reside in sys\bin)
In this document, we are concerned with BIO messages that are received as an SMS and which are displayed using the SMS UI. It is also possible that a BIO message can be opened from a file, for example an email attachment, and that they are display using an independent application.
The capabilities required to use Plugin BIO control API are LocalServices, ReadDeviceData, ReadUserData, UserEnvironment, WriteUserData WriteDeviceData, NetworkControl Location, SwEvent, and NetworkServices. Note some
of these capabilities are not user-grantable and therefore applications implementing
the Plugin BIO Control API must be granted approval to use them through the
Symbian Signed process. (Note that a BIO parser requires further capabilities.)
The most important use cases of the Plugin BIO control are the following:
If a BIO message arrives in Inbox on the device it is marked as a BIO message. This is done by the messaging framework as a result of the message arriving on a certain port or being identified by the defined character string.
Behavior on opening the message is dependent upon the values defined in
the BIF file. If the message_app_UID was KUidUseDefaultApp then
the SMS editor is opened showing the control specified by message_appctrl_name.
The framework calls NewL() on the BIO control and the control
will then be drawn. The SMS editor serves as a container for the BIO control.
As stated earlier, the BIO control itself is a polymorphic DLL; its NewL() function
should be its first (only) exported function.
The BIO control must derive from CMsgBioControl. As CMsgBioControl is
derived from the mixin MMsgBioControl, it is necessary for
the BIO control to implement any of its pure virtual functions that have not
been implemented by CMsgBioControl. To ensure that the control
is sized appropriately and that scrolling works correctly, it is also necessary
to implement various UI functions.
A header file for an example BIO control (CBioControlExample)
is shown below:
#include <msgbiocontrol.h>
class CRichBio;
class CBioControlExample : public CMsgBioControl
{
public:
IMPORT_C static CMsgBioControl* NewL(
MMsgBioControlObserver& aObserver,
CMsvSession* aSession,
TMsvId aId,
TMsgBioMode aEditorOrViewerMode,
const RFile* aFile);
// Destructor
~CBioControlExample();
public: // From MMsgBioControl
void SetAndGetSizeL(TSize& aSize);
void SetMenuCommandSetL(CEikMenuPane& aMenuPane);
TBool HandleBioCommandL(TInt aCommand);
TRect CurrentLineRect() const;
TBool IsFocusChangePossible(TMsgFocusDirection aDirection) const;
HBufC* HeaderTextL(void) const;
TInt VirtualHeight();
TInt VirtualVisibleTop();
TBool IsCursorLocation(TMsgCursorLocation aLocation) const;
protected: // From CCoeControl
TInt CountComponentControls() const;
CCoeControl* ComponentControl(TInt aIndex) const;
void SizeChanged();
void FocusChanged(TDrawNow aDrawNow);
void SetContainerWindowL(const CCoeControl& aContainer);
private: // Construction
CBioBCTest(
MMsgBioControlObserver& aObserver,
CMsvSession* aSession,
TMsvId aId,
TMsgBioMode aEditorOrViewerMode,
const RFile* aFile);
// Second phase constructor.
void ConstructL();
private:
// The viewer control
CRichBio* iViewer;
};
In the example shown, the BIO control owns a CRichBio object.
This is another CCoeControl that is designed specifically
for use with the BIO control and should be used to display the data. Many
of the functions relating to size and scrolling can then simply call the equivalent CRichBio function.
(It is possible to define another control rather than use CRichBio but
the BIO control interface requirements must be fulfilled.)
An example implementation of this class is shown below:
CBioControlExample::CBioControlExample(MMsgBioControlObserver& aObserver, CMsvSession* aSession, TMsvId aId,
TMsgBioMode aEditorOrViewerMode, const RFile* aFile):
CMsgBioControl(aObserver, aSession, aId, aEditorOrViewerMode, aFile)
{
}
CBioControlExample::~CBioControlExample()
{
delete iViewer;
}
EXPORT_C CMsgBioControl* CBioControlExample::NewL(
MMsgBioControlObserver& aObserver,
CMsvSession* aSession,
TMsvId aId,
TMsgBioMode aEditorOrViewerMode,
const RFile* aFile)
{
CBioBCTest* self = new(ELeave) CBioBCTest(
aObserver,
aSession,
aId,
aEditorOrViewerMode,
aFile);
CleanupStack::PushL(self);
self->ConstructL();
CleanupStack::Pop(self);
return self;
}
void CBioControlExample::ConstructL()
{
// Load ui resource files
_LIT(KAvkonResourceFile, "avkon.rsc");
LoadResourceL(KAvkonResourceFile);
LoadStandardBioResourceL();
}
TInt CBioControlExample::CountComponentControls() const
{
return 1; // the viewer component
}
CCoeControl* CBioControlExample::ComponentControl(TInt aIndex) const
{
if (aIndex == 0 && iViewer)
{
return iViewer;
}
return NULL;
}
void CBioControlExample::SetContainerWindowL(const CCoeControl& aContainer)
{
CCoeControl::SetContainerWindowL(aContainer);
// The reason for creating the viewer control here is that the
// construction of the viewer requires a parent with a window.
// So it cannot be done in ConstructL().
//
// The type (TRichBioMode) of CRichBio object is needed for construction
// but is not used meaningfully. Therefore either ERichBioModeEditorBase
// or ERichBioModeStandard are equivalent in terms of their effect on the CRichBio object
iViewer = CRichBio::NewL(this, ERichBioModeEditorBase);
iViewer->ActivateL();
}
void CBioControlExample::SetAndGetSizeL(TSize& aSize)
{
iViewer->SetAndGetSizeL(aSize);
SetSizeWithoutNotification(aSize);
}
void CBioControlExample::SetMenuCommandSetL(CEikMenuPane& aMenuPane)
{
}
TBool CBioControlExample::HandleBioCommandL(TInt aCommand)
{
return EFalse;
}
TInt CBioControlExample::VirtualHeight()
{
return iViewer->VirtualHeight();
}
TInt CBioControlExample::VirtualVisibleTop()
{
return iViewer->VirtualVisibleTop();
}
TBool CBioControlExample::IsCursorLocation(TMsgCursorLocation aLocation) const
{
return iViewer->IsCursorLocation(aLocation);
}
TRect CBioControlExample::CurrentLineRect() const
{
return iViewer->CurrentLineRect();
}
void CBioControlExample::SizeChanged()
{
iViewer->SetExtent(Position(), iViewer->Size());
}
void CBioControlExample::FocusChanged(TDrawNow aDrawNow)
{
iViewer->SetFocus(IsFocused());
}
TBool CBioControlExample::IsFocusChangePossible(
TMsgFocusDirection aDirection) const
{
if (aDirection == EMsgFocusUp)
{
return iViewer->IsCursorLocation(EMsgTop);
}
return EFalse;
}
HBufC* CBioControlExample::HeaderTextL() const
{
_LIT(KHeader, "My Bio control");
HBufC* buf = KHeader().AllocL();
return buf;
}
This code provides a skeleton implementation of a BIO control plug-in to the messaging UI.
When a BIO message is received by a device, its data is stored by Message
Server. If a BIO parser has been installed for the particular BIO message
type, then the message will have been parsed and maybe modified by the parser.
It is likely that any BIO control implementation requires access to the BIO
message that has been opened. Fortunately, when the BIO control constructor
is called by the framework, the ID(TMsvId) of the appropriate
message is passed through and also a handle to a session with the Message
Server. These are maintained by the base class (CMsgBioControl)
and can be accessed through iId and MSvSession() respectively.
The usual messaging APIs can be used to access the message.
In the example below, access to the data is through creation of a CMsvEntry object
loaded with the appropriate message details. Its store is then read and the
message body data passed to the viewer's (CRichBio) rich
text editor. This strategy is a bit simplistic and it is likely that the data
needs to be manipulated in some way, for example removing the BIO identifier
string, but it illustrates the basic concept.
void CBioControlExample::ReadMessageL()
{
CMsvEntry* entry = MsvSession().GetEntryL(iId);
CleanupStack::PushL(entry);
CMsvStore* store = entry->ReadStoreL();
CleanupStack::PushL(store);
// If the message has a message body then copy the
// the text to the viewer (CRichBio)
if (store->HasBodyTextL())
{
CEikRichTextEditor& ed = iViewer->Editor();
store->RestoreBodyTextL(*(ed.RichText()));
}
CleanupStack::PopAndDestroy(2, entry); // Entry & store
}
The BIO control is a CCoeControl and therefore has the
functionality of any normal control. However, it is usual for the BIO control
to create a CRichBio object (also a CCoeControl)
and use this to display the data. The CRichBio object needs
a parent window before it can be fully constructed and hence care must be
taken not to construct it until the parent window can be set. A suitable place
is in the BIO control's SetContainerWindowL() function. A
typical implementation is shown:
void CBioControlExample::SetContainerWindowL(const CCoeControl& aContainer)
{
// Call the base class
CCoeControl::SetContainerWindowL(aContainer);
// The reason for creating the viewer control here is that the
// construction of the viewer requires a parent with a window. So it
// cannot be done in ConstructL().
//
iViewer = CRichBio::NewL(this, ERichBioModeStandard); // iViewer is a CRichBio* (member data)
iViewer->ActivateL();
}
CRichBio owns an editor that is used to display rich text;
it is accessed through CRichBio::Editor(). It also has a
function, AddItemL() , that can be used to add labels to
the control. This actually appends the label and its value to the text contained
in the rich text editor. An example is shown below, where iViewer is
the CRichBio object:
CEikRichTextEditor& ed = iViewer->Editor();
if (store->HasBodyTextL())
{
store->RestoreBodyTextL(*(ed.RichText()));
}
// Define strings used for labels. Note, these should usually be defined
// in a resource file (not using _LIT).
_LIT(KLabel1, "1st label desc");
_LIT(KLabel1Text, "text for label 1");
_LIT(KLabel2, "2nd label desc");
_LIT(KLabel2Text, "this is the text for label 2");
iViewer->AddItemL(KLabel1, KLabel1Text);
iViewer->AddItemL(KLabel2, KLabel2Text);
The rich text editor is first populated with the BIO message data.
(See Retrieving data from the BIO message for
further information.) Two labels are then added immediately after the end
of the text. The displayed control (depending on the message data) would look
similar to Figure 2. (Adding a new line before the
labels is the responsibility of the developer.)

Figure 2: Example BIO control display
Menu options can be made available to the user through the left (Options) softkey. Certain menu options are available as standard on a BIO control but, if required, these can be suppressed. In addition, control specific options can be defined.
The function MMsgBioControl::OptionMenuPermissionsL() should
be implemented by the control if the standard menu options are not acceptable.
The function should return a TUint32 representing permissible
options. Options are defined by the enumeration TMsgBioOptionMenu in mmsgbiocontrol.h.
TUint32 CBioControlExample::OptionMenuPermissionsL() const
{
// Don't implement this function if the defaults are ok
// implement to define which of the standard options are presented
return (TUint32)(EMsgBioMessInfo);
}
If further options are required, they can be added to the menu through
the SetMenuCommandSetL(), which is called by the framework.
This function receives a reference to the menu pane (CEikMenuPane)
of the BIO control. CMsgBioControl::AddMenuItemL() can then
be used to insert/append options to the menu. As menu options are added, they
need to be assigned a command ID so that they can be handled when selected.
The mechanism used to do this is for the control to enquire what the next
free command ID is and then add each item as an offset against this value.
Enquiry about the next available command ID is achieved with a call to FirstFreeCommand() on
the control's observer. AddMenuItemL() then takes an offset
value as a parameter.
const TInt KItemOneCommandOffset = 0; const TInt KItemTwoCommandOffset = 1; const TInt KItemMenuPositionOne = 0; const TInt KItemMenuPositionTwo = 1;
void CBioControlExample::SetMenuCommandSetL(CEikMenuPane& aMenuPane)
{
// This is called when menu is activated
// so menu pane is passed to control and items can be added
LoadResourceL(KBCTestResourceFile); // Resource file resides in /resource directory
iMyDefinedCommand = iBioControlObserver.FirstFreeCommand();
// Add Menu items
// Value of first command added will be iMyDefinedCommand + KItemOneCommandOffset
// It will be placed at the position in the menu defined by KItemOneMenuPosition
AddMenuItemL(aMenuPane, R_MENU_ITEM_ONE, KItemOneCommandOffset, KItemOneMenuPosition);
// Value of second command added will be iMyDefinedCommand + KItemTwoCommandOffset
// It will be placed at the position in the menu defined by KItemTwoeMenuPosition
AddMenuItemL(aMenuPane, R_MENU_ITEM_TWO, KItemTwoCommandOffset, KItemTwoMenuPosition);
}
When any menu item is selected, HandleBioCommandL() is
called on the control. The command can either be dealt with by the derived
class or passed on to the base class by returning EFalse.
TBool MMsgBioControl::HandleBioCommandL(TInt aCommand)
{
TBool commandHandled = EFalse;
if (aCommand == iMyDefinedCommand + KItemOneCommandOffset)
{
// Handle the first command here
...
commandHandled = ETrue;
}
else if (aCommand == iMyDefinedCommand + KItemTwoCommandOffset)
{
// Handle the second command here
...
commandHandled = ETrue;
}
return commandHandled;
} The base class, CMsgBioControl, provides a function that
can be used to present a question to the user that has a Yes/No answer. This
is useful if you need to confirm an action with the user before carrying it
out. CMsgBioControl::ConfirmationQueryL() has two overloads
- one that takes a TDesC&, the other a resource ID of
a string. Before using the query, CMsgBioControl::LoadStandardBioResourceL() must
be called to ensure the resource is available. Any further resources should
be loaded using CMsgBioControl::LoadResourceL(). It is usual
to call this function during construction of the control.
// _LIT used for demonstration purposes only
_LIT(KQueryText, "Confirm action");
TBool response = ConfirmationQueryL(KQueryText);
if (response)
{
// Carry out action
// ...
}
The resulting query dialog is shown in Figure 3.
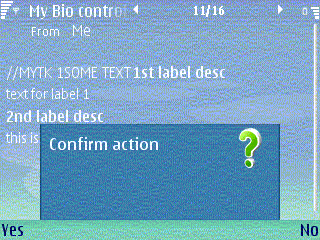
Figure 3: Example confirmation query note